Evaluating Performance
Contents
Note
Click here to download the full example code
Evaluating Performance¶
The Green’s functions based interpolations in Verde are all linear regressions
under the hood. This means that we can use some of the same tactics from
sklearn.model_selection to evaluate our interpolator’s performance. Once
we have a quantified measure of the quality of a given fitted gridder, we can
use it to tune the gridder’s parameters, like damping for a
Spline (see Model Selection).
Verde provides adaptations of common scikit-learn tools to work better with
spatial data. Let’s use these tools to evaluate the performance of a
Spline on our sample air temperature data.
import cartopy.crs as ccrs
import dask
import matplotlib.pyplot as plt
import numpy as np
import pyproj
from sklearn.model_selection import ShuffleSplit
import verde as vd
data = vd.datasets.fetch_texas_wind()
# Use Mercator projection because Spline is a Cartesian gridder
projection = pyproj.Proj(proj="merc", lat_ts=data.latitude.mean())
proj_coords = projection(data.longitude.values, data.latitude.values)
region = vd.get_region((data.longitude, data.latitude))
# For this data, we'll generate a grid with 15 arc-minute spacing
spacing = 15 / 60
Splitting the data¶
We can’t evaluate a gridder on the data that went into fitting it. The true
test of a model is if it can correctly predict data that it hasn’t seen
before. scikit-learn has the sklearn.model_selection.train_test_split
function to separate a dataset into two parts: one for fitting the model
(called training data) and a separate one for evaluating the model (called
testing data). Using it with spatial data would involve some tedious array
conversions so Verde implements verde.train_test_split which does the
same thing but takes coordinates and data arrays instead.
The split is done randomly so we specify a seed for the random number
generator to guarantee that we’ll get the same result every time we run this
example. You probably don’t want to do that for real data. We’ll keep 30% of
the data to use for testing (test_size=0.3).
train, test = vd.train_test_split(
proj_coords, data.air_temperature_c, test_size=0.3, random_state=0
)
The returned train and test variables are tuples containing
coordinates, data, and (optionally) weights arrays. Since we’re not using
weights, the third element of the tuple will be None:
print(train)
Out:
((array([ -9471409.04145469, -9242651.69892226, -9518618.69368063,
-9226352.10603726, -9196090.97662381, -9211980.21676048,
-9111443.79343314, -9285738.73749109, -9261270.26198931,
-9322307.84752338, -9449918.0091497 , -9210462.86590059,
-9437254.33247619, -9336002.17761117, -9770852.03064925,
-9286998.42499748, -9121969.81858082, -9225655.46067393,
-9784479.55912689, -9724577.60096593, -9119011.46155829,
-9366406.45333221, -9104334.19349208, -9468488.85678097,
-9512730.60889723, -9752004.43349022, -9474730.03578958,
-9295711.26358295, -9407680.3053389 , -9409483.94881389,
-9113314.23851832, -9581641.24134595, -9051780.41245142,
-9151620.1904154 , -9526253.16341599, -9370920.33356321,
-9422548.43514848, -9363887.07831951, -9926719.2733836 ,
-9263865.98169933, -10151535.32091524, -9105488.90703955,
-9399024.72527645, -9275584.89274308, -9287132.02821771,
-9254036.60191507, -9356519.81502484, -9300826.35830558,
-9088836.21992941, -9329331.55967991, -9705882.69320157,
-9544881.26957023, -9339151.39637705, -9260420.92723128,
-9092882.48888909, -9202923.82703691, -9252023.01052236,
-9250143.02235004, -9110031.41653214, -9210586.92603379,
-9192082.88001273, -9144987.74483283, -9265793.68530754,
-9038200.59940965, -9241182.06349818, -9289031.10256449,
-9405189.55958775, -9354792.51624734, -9330810.73819114,
-9321372.62498082, -9228556.55917334, -9199536.03109192,
-9278581.42211417, -9229978.47916156, -9319244.51654206,
-9297171.35591977, -9275775.75448643, -9559682.59776963,
-9197073.91460223, -9491697.64477644, -9185221.40033809,
-9486172.19730542, -9098512.91031894, -9357970.36427462,
-9348971.2330741 , -9415391.11977161, -9570580.80331691,
-9298450.12960044, -9786178.22864295, -9638164.94664898,
-9374995.23178449, -9135062.93417687, -9363782.10436067,
-9061161.26713871, -9641390.51011218, -9244388.54078699,
-9743587.43060695, -9128583.17798902, -9688294.78354872,
-9878049.52882075, -9261222.54655341, -9959308.91606633,
-9374995.23178449, -9324979.91193077, -10002090.57584577,
-9291378.70200816, -9717038.56210237, -9108943.50459486,
-9187130.01777196, -9178474.43770947, -9821430.39264604,
-9676089.17505937, -9590325.45066986, -9334752.033192 ,
-9349658.33535028, -9217667.89671332, -9192989.47329381,
-9735189.51389807, -9062249.17907602, -9332852.95884533,
-9455815.63702027, -9796236.64251933, -9443505.05457202,
-9320723.69505337, -9754266.14514927, -9765097.54908629,
-9081077.6900608 , -9042075.09280036, -9433427.55452137,
-9587615.21391383]), array([2657710.28293861, 3302077.73567252, 2899046.15133399,
2838589.68737716, 3049850.71722841, 3290767.45780106,
2931160.45084784, 3271137.71227972, 3285325.92731206,
3104893.52310543, 3247812.59589509, 3166662.55342553,
3287606.04132201, 3103141.68482877, 3197876.76156001,
3149242.24908431, 3067381.83156894, 3400585.90023853,
3243911.5995015 , 3290869.09139339, 2992634.70248073,
3169418.17605082, 3246497.10552016, 2817264.46474637,
3252930.02674656, 3200327.43647367, 3459184.29516943,
3112292.18628403, 2917869.77563751, 3197809.62807528,
3290225.42947942, 3652939.53050434, 3222645.66433888,
2963817.97546003, 2824339.57488837, 3107155.82534808,
2698337.04679593, 2601703.23660133, 3025820.22660258,
3166283.29483646, 3185632.21052805, 3023883.40574366,
3417009.35720713, 3393428.62105992, 3316162.95136998,
3082031.84084665, 2953318.41751121, 2933549.90421567,
2932676.98398158, 3020406.81467849, 3574040.01091357,
2790505.48355737, 2973418.65910265, 3307698.11129963,
2945505.02203083, 3311522.23776724, 2975313.52946633,
2861050.32840049, 2887880.3960125 , 2945898.28967202,
2941474.84700705, 3225124.76062141, 3281387.74953726,
3159592.7976058 , 3315823.31326005, 2571845.86314014,
3331844.27865595, 2740231.34771917, 3387759.04557859,
3060081.02436475, 3041845.53079302, 3210124.8374159 ,
3164074.94938398, 3271836.50108902, 2579123.45324659,
2545954.59014788, 3342747.49976745, 2979356.30060547,
3009203.6131726 , 2719110.10495866, 2846814.97160993,
3188985.42191104, 2983466.36042869, 3294675.45126596,
3039773.54716658, 3071437.56540843, 3483134.04990316,
3294607.6695262 , 3667917.1194501 , 3619924.68699415,
2573548.51452294, 2821249.74463367, 2683074.91768678,
3285461.36507139, 3652271.03785311, 3283204.30900926,
3210853.02111693, 2943844.71518428, 3230601.11578242,
3137567.17555594, 2777047.34270749, 2945144.53916802,
3066497.95513723, 3110760.99744545, 3188091.12461515,
3298991.83921469, 3394820.85278175, 3391739.95155904,
3286590.08456857, 3389059.21988513, 3085628.36101872,
3630202.83368938, 3134419.72201146, 3256654.45390054,
2573738.88783378, 3339785.07776942, 3063935.13249589,
3648601.05705207, 3330211.5352512 , 3142162.17467495,
2982622.30031315, 3282673.97488324, 3183676.67838886,
3006522.74236789, 2990221.2234946 , 3532083.29147308,
2942206.50247381, 3121339.67312344, 2913261.59698675,
3259243.23756799])), (array([18.15166667, 12.17857143, 16.45916667, 16.60084507, 12.41170213,
9.67159091, 16.33043478, 9.43978723, 11.10313725, 10.22955556,
11.57575758, 12.47194444, 12.22138889, 10.36794118, 8.775 ,
10.46055556, 12.55241379, 11.78611111, 7.89736111, 7.06944444,
14.57882353, 12.33319444, 11.96138889, 16.88692308, 10.20833333,
9.1525 , 9.64888889, 10.29677419, 15.16444444, 12.81565217,
10. , 3.16277778, 13.49071429, 14.98529412, 16.66666667,
13.06835821, 16.63416667, 20.43375 , 5.96285714, 13.76470588,
9.16391304, 13.8736 , 10.99888889, 11.23633803, 9.751875 ,
12.43888889, 10.94418605, 15.23805556, 16.66589744, 11.40892857,
4.984 , 18.08955224, 11.45891304, 12.05545455, 16.13939394,
11.18275362, 14.49676056, 15.90777778, 18.8625 , 14.27527778,
14.97291667, 10.73239437, 8.98125 , 11.89032258, 11.46025641,
22.6082 , 10.77111111, 16.40823529, 9.23611111, 10.82863636,
12.57958333, 11.18545455, 11.13193548, 10.56277778, 20.97333333,
23.26113208, 10.365 , 14.46875 , 14.21430556, 16.8525 ,
17.01530612, 12.83722222, 15.22228571, 9.005 , 14.1 ,
14.19444444, 9.02333333, 12.11923077, 2.1525 , 5.64638889,
21.09483871, 19.73611111, 18.09676056, 11.48449275, 4.29166667,
9.78117647, 10.1875 , 16.36536585, 8.73277778, 7.70694444,
17.2974 , 15.35777778, 14.53130435, 16.58333333, 7.52666667,
11.37028571, 5.79375 , 8.54088235, 9.196 , 10.37263889,
11.39375 , 7.08166667, 11.74291667, 10.86027778, 20.47454545,
10.9936 , 12.47652778, 3.43633803, 10.0825 , 9.59677419,
13.55277778, 6.09319444, 20.58333333, 11.65820513, 14.4926087 ,
3.14875 , 16.80952381, 12.45724138, 15.76208333, 11.69902778]),), (None,))
print(test)
Out:
((array([ -9355174.23973406, -9305731.50511056, -9378946.06987249,
-9894654.50049513, -9357159.20186525, -9310436.24708494,
-9176098.20900436, -9848465.95859632, -9657890.50782771,
-9408987.70828107, -9592090.92179618, -9397058.84931957,
-9614660.3229513 , -9438991.174341 , -9220530.82286406,
-9464280.35533937, -9148327.82534205, -9254885.93667311,
-9284574.48085645, -9103675.72047743, -9617332.38735867,
-9038401.00424027, -9631570.67341502, -9133335.63539924,
-9125099.95117223, -9369183.49169843, -8985427.32736408,
-9520794.51755526, -9397182.90945276, -8972410.55646536,
-9418511.70927591, -9312192.17512406, -9633794.21272551,
-9386466.02256181, -9298192.46624686, -10151936.13057628,
-9335849.48821648, -9334064.93091582, -8951759.31583123,
-9066696.25769684, -9086927.60249552, -9399253.75936852,
-9167595.31833663, -9052610.66103518, -9401849.47907855,
-9332280.37361521, -8991029.11953241, -8973832.47645352,
-9529249.69278712, -9266070.43483542, -9429505.34569483,
-8999875.56133822, -9322689.57101016, -9604983.63256177,
-9104467.79671247, -9706970.60513881]), array([2719870.27460582, 2743685.56324252, 2933768.14517002,
3027361.13101297, 2757426.84021469, 3028296.78889616,
2903724.5557224 , 3182157.20869187, 3066100.23492454,
3255225.28091213, 2860584.19616269, 2934171.90242784,
3714012.76916745, 3011874.16439619, 3080991.84629854,
2915167.81938923, 2873187.46869299, 3003831.55941359,
2735051.95799933, 2953788.49303643, 2915178.71303919,
3249859.25595487, 2916780.19714376, 3182123.69352164,
3337515.62533228, 3230904.19741823, 3187923.45258655,
3041327.49695635, 2912727.91396922, 2979696.00616845,
2954947.37073634, 3348209.31923159, 3284716.48014213,
3043124.19066597, 3246452.13452785, 3189924.51834186,
3339251.70946121, 2715192.2711302 , 2992766.36237038,
3024323.56139652, 3192216.72345509, 3430837.16378734,
3327060.19688609, 2904921.67586422, 2871114.86619099,
2807032.29570529, 2992876.08017689, 3082728.91614068,
3254021.33225593, 3216131.11265388, 3253177.52895142,
3265222.50231348, 2744264.94274218, 3049585.98626738,
3129450.24999779, 3452568.99011507])), (array([17.71611111, 17.3918 , 10.73421875, 8.08555556, 16.258125 ,
16.18181818, 15.87828571, 7.47625 , 11.02857143, 12.25472222,
17.16923077, 15.60208333, 5.04152778, 13.14263889, 12.47835821,
16.69304348, 18.73916667, 11.46194444, 18.38255319, 16.02173913,
16.57884615, 10.55866667, 16.48916667, 13.43583333, 10.67041667,
11.90319444, 11.49557143, 12.17826087, 14.27805556, 17.43382353,
12.95833333, 11.04708333, 9.60486111, 13.46 , 10.58730159,
7.39333333, 10.11722222, 17.7375 , 17.10013889, 14.64180556,
12.44957143, 9.68391304, 11.93055556, 20.34757576, 15.4025 ,
16.76319444, 17.33347222, 13.55152778, 10.49541667, 11.92041667,
10.42583333, 12.69078125, 17.22138889, 10.33333333, 12.80555556,
3.85638889]),), (None,))
Let’s plot these two datasets with different colors:
plt.figure(figsize=(8, 6))
ax = plt.axes()
ax.set_title("Air temperature measurements for Texas")
ax.plot(train[0][0], train[0][1], ".r", label="train")
ax.plot(test[0][0], test[0][1], ".b", label="test")
ax.legend()
ax.set_aspect("equal")
plt.show()
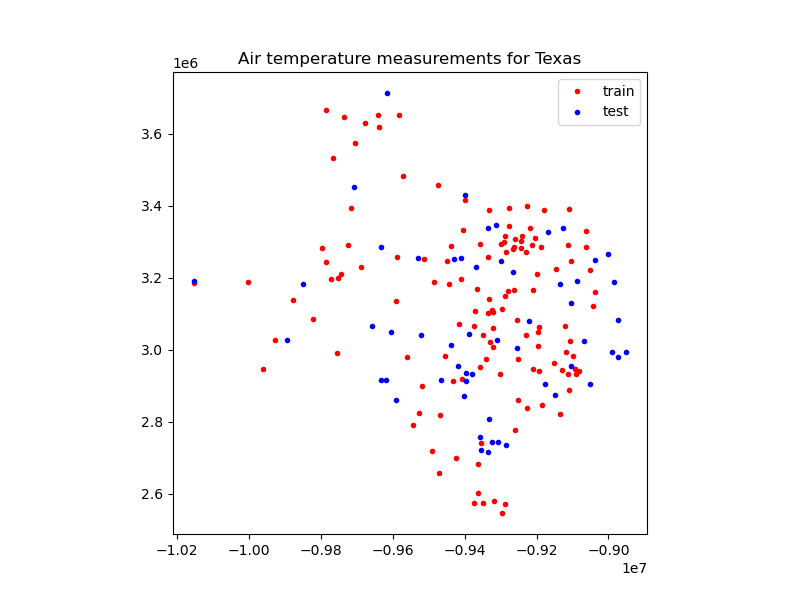
We can pass the training dataset to the fit
method of most gridders using Python’s argument expansion using the *
symbol.
spline = vd.Spline()
spline.fit(*train)
Out:
Spline()
Let’s plot the gridded result to see what it looks like. First, we’ll create a geographic grid:
Out:
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:463: FutureWarning: The 'spacing', 'shape' and 'region' arguments will be removed in Verde v2.0.0. Please use the 'verde.grid_coordinates' function to define grid coordinates and pass them as the 'coordinates' argument.
warnings.warn(
<xarray.Dataset>
Dimensions: (latitude: 43, longitude: 51)
Coordinates:
* longitude (longitude) float64 -106.4 -106.1 -105.9 ... -94.06 -93.8
* latitude (latitude) float64 25.91 26.16 26.41 ... 35.91 36.16 36.41
Data variables:
temperature (latitude, longitude) float64 36.72 36.32 35.92 ... 4.568 4.711
Attributes:
metadata: Generated by Spline()
Then, we’ll mask out grid points that are too far from any given data point and plot the grid:
mask = vd.distance_mask(
(data.longitude, data.latitude),
maxdist=3 * spacing * 111e3,
coordinates=vd.grid_coordinates(region, spacing=spacing),
projection=projection,
)
grid = grid.where(mask)
plt.figure(figsize=(8, 6))
ax = plt.axes(projection=ccrs.Mercator())
ax.set_title("Gridded temperature")
pc = grid.temperature.plot.pcolormesh(
ax=ax,
cmap="plasma",
transform=ccrs.PlateCarree(),
add_colorbar=False,
add_labels=False,
)
plt.colorbar(pc).set_label("C")
ax.plot(data.longitude, data.latitude, ".k", markersize=1, transform=ccrs.PlateCarree())
vd.datasets.setup_texas_wind_map(ax)
plt.show()
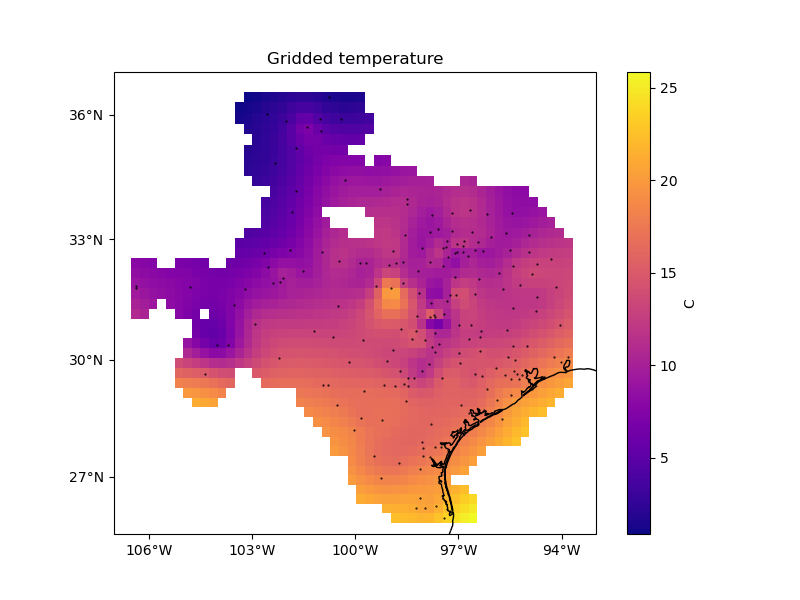
Scoring¶
Gridders in Verde implement the score method
that calculates the R² coefficient of determination for a given
comparison dataset (test in our case). The R² score is at most 1, meaning
a perfect prediction, but has no lower bound.
score = spline.score(*test)
print("R² score:", score)
Out:
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
R² score: 0.8404552994992693
That’s a good score meaning that our gridder is able to accurately predict data that wasn’t used in the gridding algorithm.
Caution
Once caveat for this score is that it is highly dependent on the
particular split that we made. Changing the random number generator seed
in verde.train_test_split will result in a different score.
# Use 1 as a seed instead of 0
train_other, test_other = vd.train_test_split(
proj_coords, data.air_temperature_c, test_size=0.3, random_state=1
)
print("R² score with seed 1:", vd.Spline().fit(*train_other).score(*test_other))
Out:
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
R² score with seed 1: 0.7832246926768177
Cross-validation¶
A more robust way of scoring the gridders is to use function
verde.cross_val_score, which (by default) uses a k-fold
cross-validation
by default. It will split the data k times and return the score on each
fold. We can then take a mean of these scores.
scores = vd.cross_val_score(vd.Spline(), proj_coords, data.air_temperature_c)
print("k-fold scores:", scores)
print("Mean score:", np.mean(scores))
Out:
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
k-fold scores: [0.81655898 0.72019255 0.81596348 0.87740087 0.75006854]
Mean score: 0.7960368854873764
You can also use most cross-validation splitter classes from
sklearn.model_selection by specifying the cv argument. For
example, if we want to shuffle then split the data n times
(sklearn.model_selection.ShuffleSplit):
shuffle = ShuffleSplit(n_splits=10, test_size=0.3, random_state=0)
scores = vd.cross_val_score(
vd.Spline(), proj_coords, data.air_temperature_c, cv=shuffle
)
print("shuffle scores:", scores)
print("Mean score:", np.mean(scores))
Out:
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
shuffle scores: [0.8404553 0.79219264 0.60098099 0.73123421 0.67325199 0.70560489
0.68905622 0.83517217 0.86908658 0.84133251]
Mean score: 0.7578367516969872
Parallel cross-validation¶
Cross-validation involves running several model fit and score operations which are independent of each other. Because of this, they are prime targets for parallelization. Verde uses the excellent Dask library for parallel execution.
To run verde.cross_val_score with Dask, use the delayed argument:
scores = vd.cross_val_score(
vd.Spline(), proj_coords, data.air_temperature_c, delayed=True
)
print("Delayed k-fold scores:", scores)
Out:
Delayed k-fold scores: [Delayed('fit_score-e8287a90-5a56-4b29-9e76-e963e3c7fae5'), Delayed('fit_score-f71100f2-3479-41e2-8a9a-65ffebc1011d'), Delayed('fit_score-a783cca6-c875-4302-879c-7edef87b4b5d'), Delayed('fit_score-e8965c27-6ec5-4a61-b5ed-3cc0ec58b9ea'), Delayed('fit_score-4a3ec24a-ab06-4571-9a01-925c21cfbf60')]
In this case, the scores haven’t actually been computed yet (hence the
“delayed” term). Instead, Verde scheduled the operations with Dask. Since we
are interested only in the mean score, we can schedule the mean as well using
dask.delayed:
mean_score = dask.delayed(np.mean)(scores)
print("Delayed mean:", mean_score)
Out:
Delayed mean: Delayed('mean-4a682b29-9d05-4142-a818-8b86a0ce0338')
To run the scheduled computations and get the mean score, use
dask.compute or .compute(). Dask will automatically execute
things in parallel.
mean_score = mean_score.compute()
print("Mean score:", mean_score)
Out:
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
/usr/share/miniconda3/envs/test/lib/python3.9/site-packages/verde/base/base_classes.py:359: FutureWarning: The default scoring will change from R² to negative root mean squared error (RMSE) in Verde 2.0.0. This may change model selection results slightly.
warnings.warn(
Mean score: 0.7960368854873764
Note
Dask will run many fit operations in parallel, which can be memory
intensive. Make sure you have enough RAM to run multiple fits.
Improving the score¶
That score is not bad but it could be better. The default arguments for
Spline aren’t optimal for this dataset. We could try
different combinations manually until we get a good score. A better way is to
do this automatically. In Model Selection we’ll go over how to do just
that.
Total running time of the script: ( 0 minutes 0.456 seconds)
